Kaggle plant pathology write-up
- Context
- Problem description
- My score evolution
- What worked
- What was complicated
- Other things I tried
- How I worked
- Some details
- Paper to cite
Context
This page is my write-up for the plant pathology kaggle competition
I worked on this for 4 weeks as part of the FastChai and Kaggle: Group based Projects organized by Sanyam Bhutani. Each sprint (each sprint is 2 weeks long), small groups work on past kaggle competitions.
For this one, I worked 2 sprints and reached top 12%. I’m happy with the result!
To be short, here’s what worked well for me: 5-fold training, pseudo-labelling augmentation, se-resnext50 pretrained models, 320x512 images, TTA, augmentation (rotate, zoom, flip, contrast, brightness), LabelSmoothingCrossEntropy loss function, managing duplicates.
I worked with Isaac Flath on this project - see his blog here
Problem description
The goal of this competition is to classify plant images. There are 4 possible output: healthy, scab , rust and multiple diseases
This is an image classification problem. Each image can only have 1 label. The category ‘multiple diseases’ is largely under-represented.
There are 1821 train images, 1821 test images. Images are quite large (2048x1365)
Some examples:
| class | example |
|---|---|
| rust |   |
| scab |   |
| healthy |  |
| multiple_diseases |  |
My score evolution
I worked 2 * 2 weeks on this competition.
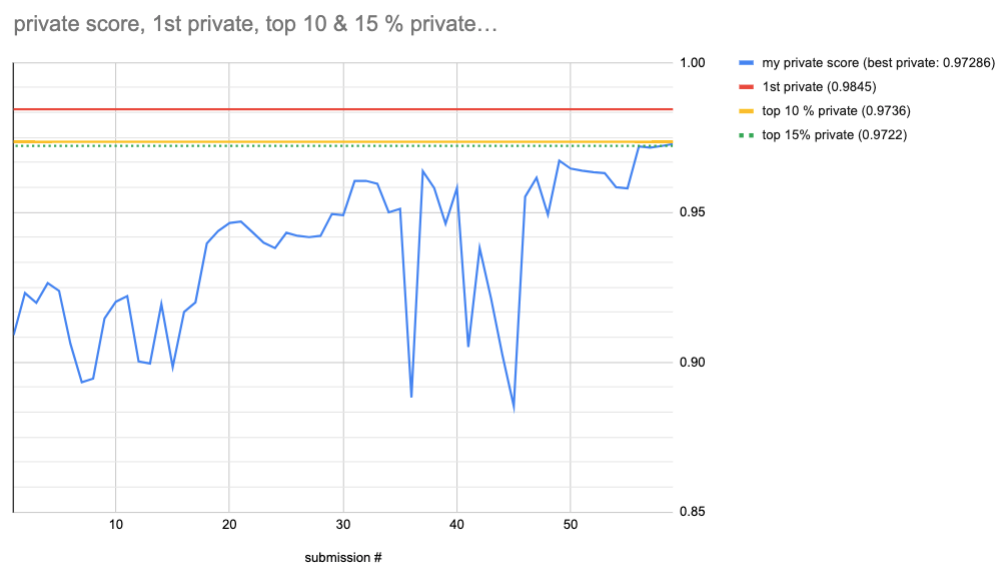
Final best score: 0.97286 private. That would give me rank #160 (out of 1317). Top 12%…
Frist 2 weeks: My best score is: 0.96380 private, 0.97800 public. That would give me rank #406 (out of 1317). Top 33%…
Top 10% threshold is 0.9736 private, 1st place solution achieved 0.98445 private.
I mostly looked at the 1st place solution for inspiration discussion on kaggle
What worked
- seresnextnet50 as architecture
- larger images (320x512). I used batch size 16 to train on kaggle notebooks (bs=64 gives OOM)
- 5-fold stratified cross-validation. It clearly helps because when I submit the 5 scores from the folds, they perform worse.
- Instead of using the 5 folds, I used the best 4 (according to public LB score) and it gave me a bump of 0.00064 points. It sounds like nothing but that improved my rank by 40 positions from 200 to 160.
- augmentations (Rotate, Zoom, Warp, Brightness, Flip, Contrast, Resize, Normalize
- TTA
- LabelSmoothingCrossEntropy loss function
- pseudo labeling technique to augment the dataset (see Isaac’s blog about pseudo labeling)
- generate 5 predictions from the 5-fold cross-validation and use rankdata to blend the submissions
- I adapted a notebook to detect duplicate images. See my version here Forked from Appian notebook here
This helped me see that many images were duplicated (either both images were in the training set or test set or 1 image in train and the other one in test).
What was disappointing is that for images present in train and test datasets, I used the train label to change my predictions… and that made my score worse… So it means that the labels are not necessarily consistent…
There’s actually 1 duplicated image in the training with 2 different labels…

Also based on this notebook, I removed duplicated images in the train set.
What was complicated
- some images are mislabeled -> what to do with these? ignore? change the label?
- some noisy labels as well (rust in the background of the healthy leaf)

- hard to manually label data when mislabeled - especially for the ‘multi-disease’ category
- ROC AUC metric on validation set is >0.999 on all folds -> hard to know what is helping before submitting
Other things I tried
- use soft labels and try to apply distilation knowledge
- use gradcam to visualize and interpret results
- use bigger images. I saved the models trained with images 320x512 and used the models to train 500x750 images. For that, I had to decrease again the batch size (bs=8) to avoid out of memory. It didn’t help much.

- oversampling the under-represented class (‘multiple_disease’). This seemed to help the local score but didn’t translate on the leaderboard.
How I worked
- Kaggle kernels
- 5-fold cross validation in 1 notebook
Some details
Use seresnextnet50 as architecture
!pip install -q pretrainedmodels
import pretrainedmodels
def model_f(pretrained=True,**kwargs):
return pretrainedmodels.se_resnext50_32x4d(num_classes=1000,pretrained='imagenet')
learn = cnn_learner(dls, model_f, pretrained=True,
loss_func=LabelSmoothingCrossEntropy(), metrics=[RocAuc(), error_rate])
Use rankdata to blend the fold submissions
When running the cross validation, you generate a prediction of the test set for each fold. Since the metric is AUC ROC, the only thing that matters is the order of the predictions. Rankdata worked better than averaging the predictions.
An idea I got at the end was to ignore the worst prediction from my blending. It paid off because my score went from .97220 to .97286 (looks small but that going from #200 to #160)
Here are my fold scores:
| Fold | My CV ROC AUC | My CV ROC AUC (multiple diseases) | Private score | Public score |
|---|---|---|---|---|
| 0 | .9835 | .94660 | .97062 | .96879 |
| 1 | .98645 | .96257 | .9633 | .96487 |
| 2 | .98934 | .96798 | .96172 | .96011 |
| 3 | .99286 | .9780 | .96414 | .97340 |
| 4 | .98576 | .96202 | .96855 | .97126 |
Ignoring the fold # 2 helped!
Run a stratified K-fold cross-validation
for fold in range(0,5):
plant = DataBlock(blocks=(ImageBlock, CategoryBlock),
splitter=IndexSplitter(train[train['fold'] == fold].index),
get_x=ColReader('image_id', pref=path/"images", suff='.jpg'),
get_y=ColReader('label'),
item_tfms=Resize(R2),
batch_tfms=comp)
dls = plant.dataloaders(train, bs=16)
learn = cnn_learner(dls, model_f, pretrained=True, loss_func=LabelSmoothingCrossEntropy(), metrics=[RocAuc(), error_rate])
print(f'-------- {fold} ---------')
learn.fine_tune(10, base_lr=5e-2)
learn.unfreeze()
learn.fit_one_cycle(15, lr_max=slice(1e-7, 1e-4))
learn.save(f'seresnext50-fold-{fold}')
# prediction test set
tst_dl = dls.test_dl(test)
preds, _ = learn.tta(dl=tst_dl)
cols=['image_id'] + list(dls.vocab)
res = pd.concat([test, pd.DataFrame(preds,columns = learn.dls.vocab)],axis=1)[cols]
res.to_csv(f'submission_tta_resnext_{fold}.csv',index=False)
# prediction valiation set
res = learn.get_preds()
cols= dls.vocab
r = pd.DataFrame(res[0], columns=cols)
valid_with_preds = pd.concat([train[train['fold'] == fold].reset_index(), r], axis=1)
# print metric for each column
print(roc_auc_score(train[train['fold'] == fold][cols], r))
for col in cols:
score = roc_auc_score(train[train['fold'] == fold][col], r[col])
print(f'{col} -> {score}')
valid_with_preds.to_csv(f'validation_resnext_{fold}.csv',index=False)
Pseudo-labeling as data augmentation technique
The idea is to use the test dataset at training time.
With the trained model, you run inference on the test data. For the images where you are confident, you integrate these images at training time.
The idea behind is that you might learn more about the structure of the images thanks to these additional images.
I tried this and it was a bit tedious because I manually reviewed the predictions before using them at training time.
Use soft labels
In the 1st place solution, they used knowledge distillation and soft labels.
Thanks to my teammate Isaac Flath, I was able to implement a callback to do something similar.
This does not implement what the 1st place solution did exactly but I wanted to try this way as it sounded interested to try and code.
I trained 5-fold models and stored the validation prediction for each fold. That gave me OOF predictions for the entire training set.
I then used this to generate soft labels: 0.7 * ground truth + 0.3 * model prediction For example, if the truth is (0,1,0,0) and the prediction is (0,0,1,0), the label I used is (0,0.7,0.3,0). This is similar to a MixUp augmentation technique.
I used Logsoftmax in the loss function.
In this code, train_with_preds is an dataframe with the soft labels for the whole entire training set.
class MyMixUp(Callback):
def __init__(self, train_with_preds):
# training data with soft predictions (0, 0.3, 0.7, 0)
self.train_with_preds = train_with_preds
# dictionary to help decode
self.d={x:idx for idx,x in enumerate(['healthy', 'multiple_diseases', 'rust', 'scab'])}
# columns with the soft values
self.soft=[f'new_{x}' for x in ['healthy', 'multiple_diseases', 'rust', 'scab']]
def before_batch(self):
# get the idx from the dataloader
self.idxs = self.dl._DataLoader__idxs
# get the images' names for the current batch
imgs = L(o for o in self.dl.items.iloc[self.idxs[self.iter*self.dl.bs:self.iter*self.dl.bs+self.dl.bs]].image_id)
# get the soft label corresponding to each image
df = self.train_with_preds.set_index('image_id').loc[imgs, self.soft]
soft_labels = [tuple(x) for x in df.to_records(index=False)]
# replace the targets with the soft targets
self.learn.yb = (Tensor(soft_labels).cuda(),)
def MyLoss(output, target):
return torch.mean(torch.sum(-target * nn.LogSoftmax(dim=-1)(output), -1))
learn1 = cnn_learner(dls, model_f, pretrained=True,
loss_func=MyLoss, metrics=[mse, exp_rmspe],
cbs=MyMixUp(train_with_preds))
I am sure there are much better ways to do this but I was pretty happy to be able to modify the training loop this way.